Exam 15: Genomics and Proteomics
Exam 1: Introduction to Genetics69 Questions
Exam 2: Chromosomes and Cellular Reproduction70 Questions
Exam 3: Basic Principles of Heredity65 Questions
Exam 4: Sex Determination and Sex-Linked Characteristics131 Questions
Exam 5: Linkage, Recombination, and Eukaryotic Gene Mapping60 Questions
Exam 6: Chromosome Variation49 Questions
Exam 7: Bacterial and Viral Genetic Systems57 Questions
Exam 8: DNA- the Chemical Nature of the Gene87 Questions
Exam 9: DNA Replication and Recombination59 Questions
Exam 10: From DNA to Proteins- Transcription and Rna Processing50 Questions
Exam 11: From DNA to Protein- Translation58 Questions
Exam 12: Control of Gene Expression89 Questions
Exam 13: Gene Mutations, Transposable Elements, and Dna Repair76 Questions
Exam 14: Molecular Genetic Analysis and Biotechnology69 Questions
Exam 15: Genomics and Proteomics50 Questions
Exam 16: Cancer Genetics60 Questions
Exam 17: Quantitative Genetics56 Questions
Exam 18: Population and Evolutionary Genetics58 Questions
Select questions type
A linear DNA fragment is cut with a restriction enzyme to yield two fragments.How many restriction sites are there for the enzyme in this fragment?
(Multiple Choice)
4.8/5  (39)
(39)
In the following list of cloned fragments, the fragments needed to make the longest possible contig with the least amount of overlap are: 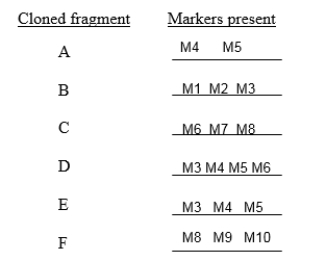
(Multiple Choice)
4.9/5  (40)
(40)
Two unrelated individuals are 99.9% similar.Since the human genome consists of roughly 3 billion bases, these two individuals will have _____ bases that are different.
(Multiple Choice)
5.0/5  (40)
(40)
How has the study of microbiomics shed light on the understanding of obesity?
(Essay)
4.8/5  (32)
(32)
A human gene with a disease phenotype is going to be mapped by positional cloning.Which would be the MOST useful for this task?
(Multiple Choice)
4.8/5  (43)
(43)
Describe one major difference in the organization or content of prokaryotic and eukaryotic genomes.
(Essay)
4.9/5  (33)
(33)
As set of overlapping DNA fragments that form a contiguous stretch of DNA is called a:
(Multiple Choice)
4.7/5  (42)
(42)
You are attempting to generate a physical map of the bacteriophage genome using restriction enzyme digestions.You digest the phage DNA with EcoRI or NcoI or both, and observe the fragment sizes listed below.You know that bacteriophage has a linear, not circular, genome. 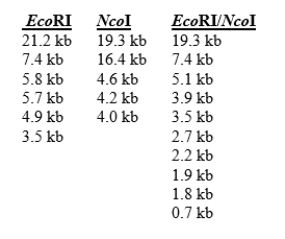 How many times does the EcoRI enzyme cut the DNA? How many times does the NcoI enzyme cut the DNA?
How many times does the EcoRI enzyme cut the DNA? How many times does the NcoI enzyme cut the DNA?
(Essay)
4.8/5  (31)
(31)
What is the main difference between map-based sequencing and whole genome shotgun sequencing?
(Essay)
5.0/5  (47)
(47)
Compare the fields of structural, functional, and comparative genomics.What is the purpose of each?
(Essay)
4.7/5  (44)
(44)
The following microarray data are shown for four human genes.Two samples were hybridized to the microarray in this experiment.One sample was cDNA generated from mRNA of a tissue culture cell line that was originally derived from a B-cell leukemia.The other sample was cDNA generated from normal, mature neural tissue. - Vertical stripes mean hybridization to cancerous B-cell cDNA only.
- Horizontal stripes mean hybridization to normal neural cell cDNA only.
- Completely filled in means hybridization to both cDNA types.
- Blank means hybridization to neither cell's cDNA. 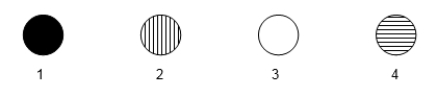 The gene sequences contained in spot 2 are present only in the B-cell cancer sample.What is a possible function for this gene, based on the microarray data?
The gene sequences contained in spot 2 are present only in the B-cell cancer sample.What is a possible function for this gene, based on the microarray data?
(Multiple Choice)
4.9/5  (26)
(26)
Which technique would NOT be used to find a gene for a functional protein in a sequenced region of a genome?
(Multiple Choice)
4.8/5  (35)
(35)
The following microarray data are shown for four human genes.Two samples were hybridized to the microarray in this experiment.One sample was cDNA generated from mRNA of a tissue culture cell line that was originally derived from a B-cell leukemia.The other sample was cDNA generated from normal, mature neural tissue. - Vertical stripes mean hybridization to cancerous B-cell cDNA only.
- Horizontal stripes mean hybridization to normal neural cell cDNA only.
- Completely filled in means hybridization to both cDNA types.
- Blank means hybridization to neither cell's cDNA. 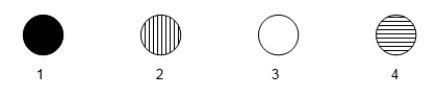 One spot on the microarray contains sequences unique to a gene for an RNA polymerase subunit.One spot contains sequences unique to the skeletal muscle myosin gene.Which spot would be expected to contain the RNA polymerase gene sequences?
One spot on the microarray contains sequences unique to a gene for an RNA polymerase subunit.One spot contains sequences unique to the skeletal muscle myosin gene.Which spot would be expected to contain the RNA polymerase gene sequences?
(Multiple Choice)
5.0/5  (35)
(35)
Which of the following statements about eukaryotic genomes is TRUE?
(Multiple Choice)
4.9/5  (32)
(32)
The transcription factor Twist helps specify development of the mesoderm in the Drosophila embryo.You wish to identify other genes that Twist (as a transcription factor)switches on or off.You have wild-type embryos available, as well as embryos that are mutant for the Twist gene.Design an experiment using microarrays that could allow you to identify genes switched on or off by the Twist transcription factor.
(Essay)
4.8/5  (32)
(32)
Showing 21 - 40 of 50
Filters
- Essay(0)
- Multiple Choice(0)
- Short Answer(0)
- True False(0)
- Matching(0)